# load packages
library(countdown)
library(tidyverse)
library(janitor)
library(colorspace)
library(broom)
library(fs)
# set theme for ggplot2
ggplot2::theme_set(ggplot2::theme_minimal(base_size = 14))
# set width of code output
options(width = 65)
# set figure parameters for knitr
knitr::opts_chunk$set(
fig.width = 7, # 7" width
fig.asp = 0.618, # the golden ratio
fig.retina = 3, # dpi multiplier for displaying HTML output on retina
fig.align = "center", # center align figures
dpi = 300 # higher dpi, sharper image
)Visualizing time series data II
Lecture 7
University of Arizona
INFO 526 - Summer 2024
Setup
Working with dates
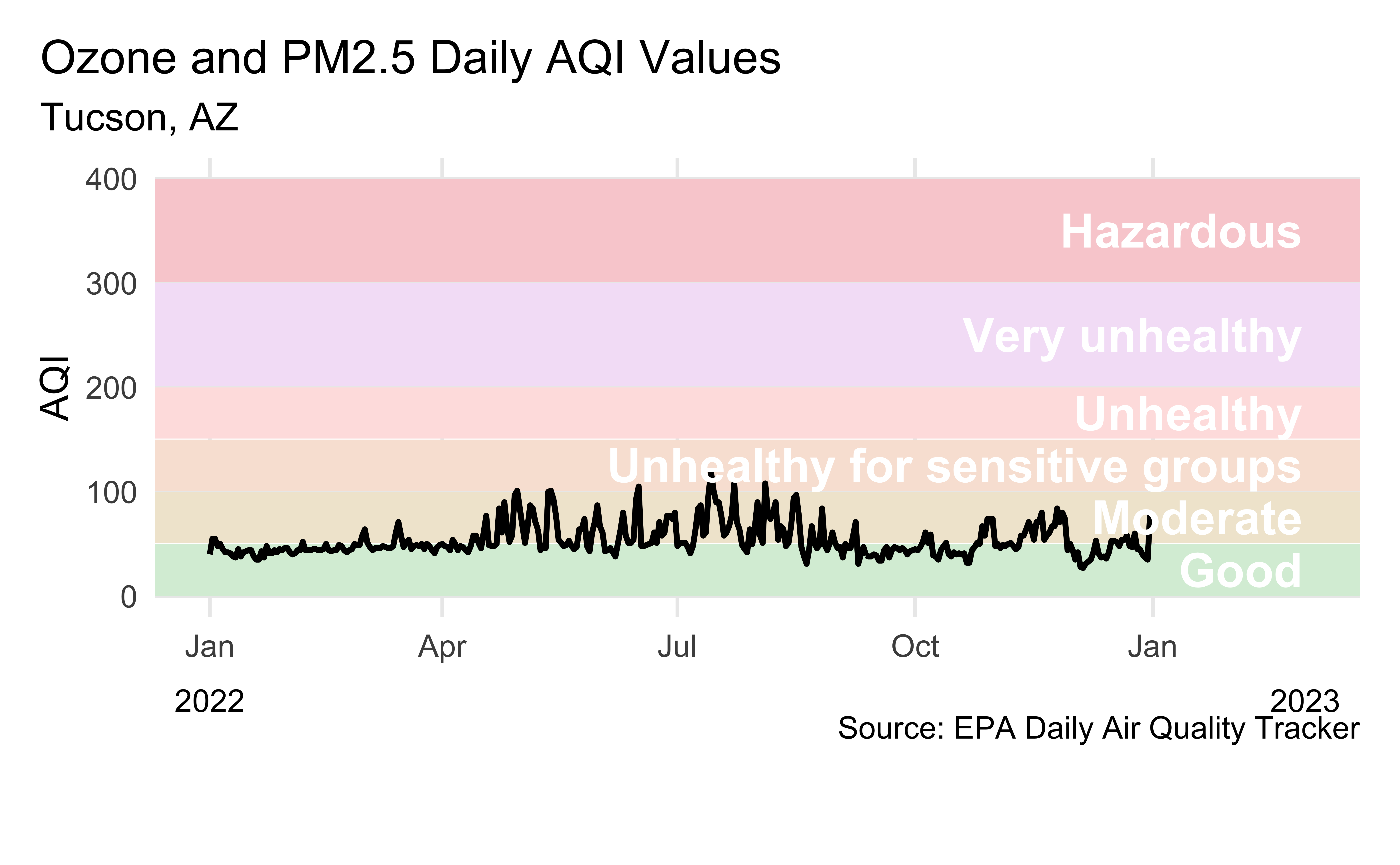
AQI levels
The previous graphic in tibble form, to be used later…
AQI data
Source: EPA’s Daily Air Quality Tracker
2016 - 2022 AQI (Ozone and PM2.5 combined) for Tucson, AZ core-based statistical area (CBSA), one file per year
2016 - 2022 AQI (Ozone and PM2.5 combined) for San Francisco-Oakland-Hayward, CA CBSA, one file per year
2022 Tucson, AZ
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source
<date> <dbl> <chr> <chr> <chr> <chr>
1 2022-01-01 40 PM2.5 GERONIMO 04-019-11… AQS
2 2022-01-02 55 PM2.5 GERONIMO 04-019-11… AQS
3 2022-01-03 55 PM2.5 GERONIMO 04-019-11… AQS
# ℹ 362 more rows
# ℹ 5 more variables: x20_year_high_2000_2019 <dbl>,
# x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>Visualizing Tucson AQI
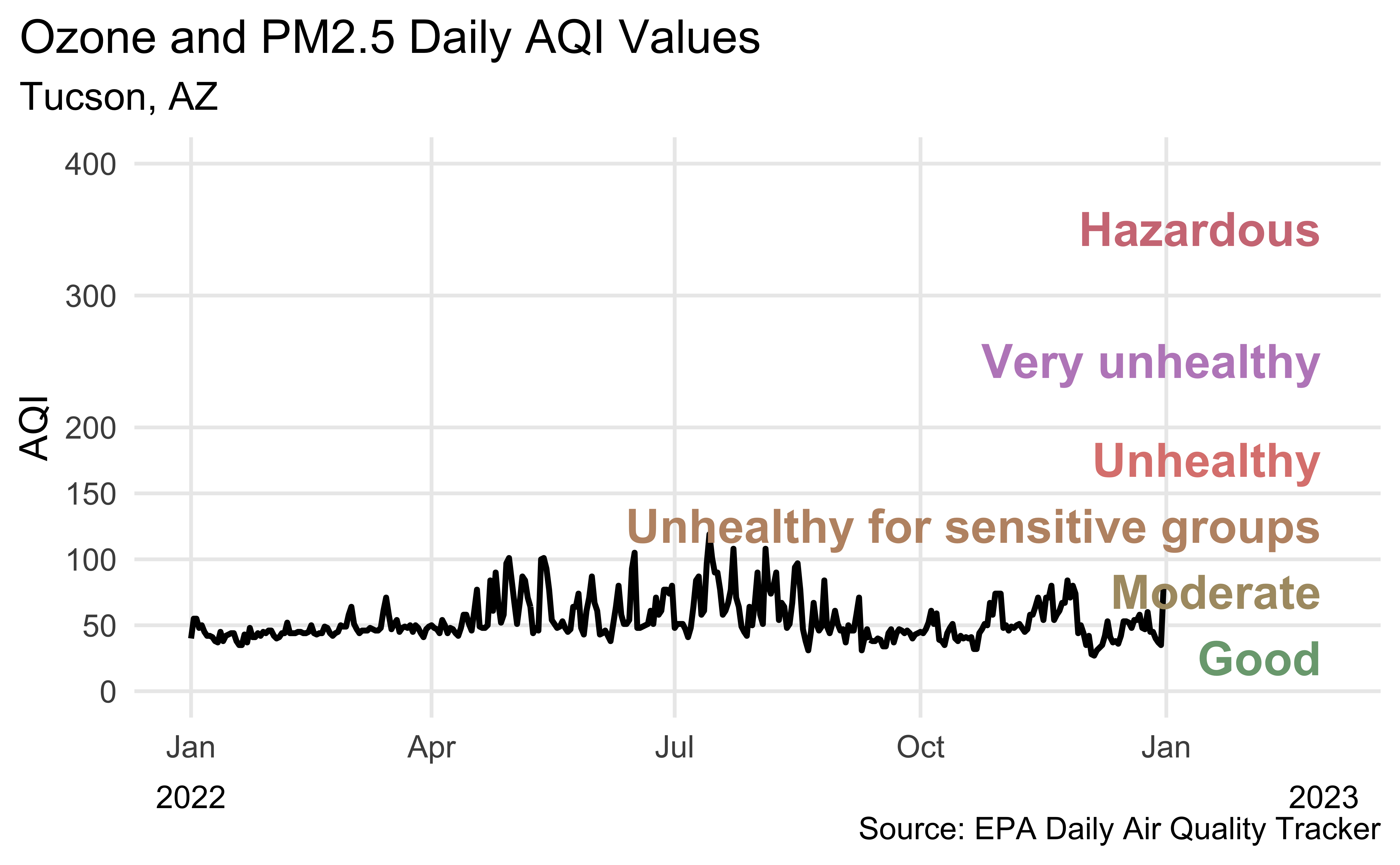
Another visualization of Tucson AQI
Recreate the following visualization.
Highlights
The lubridate package is useful for converting to dates from character strings in a given format, e.g.
mdy(),ymd(), etc.The colorspace package is useful for programmatically darkening / lightening colors
scale_x_date: Setdate_labelsas"%b %y"for month-2 digit year,"%D"for date format such as%m/%d/%y, etc. See help forstrptime()for more.scale_color_identity()orscale_fill_identity()can be useful when your data already represents aesthetic values that ggplot2 can handle directly. By default doesn’t produce a legend.
Calculating cumulatives
Cumulatives over time
When visualizing time series data, a somewhat common task is to calculate cumulatives over time and plot them
In our example we’ll calculate the number of days with “good” AQI (≤ 50) and plot that value on the y-axis and the date on the x-axis
Calculating cumulatives
Step 1. Arrange your data
Calculating cumulatives
Step 2. Identify good days
tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(good_aqi = if_else(aqi_value <= 50, 1, 0))# A tibble: 365 × 3
date aqi_value good_aqi
<date> <dbl> <dbl>
1 2022-01-01 40 1
2 2022-01-02 55 0
3 2022-01-03 55 0
4 2022-01-04 48 1
5 2022-01-05 50 1
# ℹ 360 more rowsCalculating cumulatives
Step 3. Sum over time
tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(
good_aqi = if_else(aqi_value <= 50, 1, 0),
cumsum_good_aqi = cumsum(good_aqi)
)# A tibble: 365 × 4
date aqi_value good_aqi cumsum_good_aqi
<date> <dbl> <dbl> <dbl>
1 2022-01-01 40 1 1
2 2022-01-02 55 0 1
3 2022-01-03 55 0 1
4 2022-01-04 48 1 2
5 2022-01-05 50 1 3
# ℹ 360 more rowsPlotting cumulatives

tuc_2022 |>
select(date, aqi_value) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(
good_aqi = if_else(aqi_value <= 50, 1, 0),
cumsum_good_aqi = cumsum(good_aqi)
) |>
ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) +
geom_line() +
scale_x_date(date_labels = "%b %Y") +
labs(
x = NULL, y = "Number of days",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "Tucson, AZ",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Detrending
Detrending
Detrending is removing prominent long-term trend in time series to specifically highlight any notable deviations
Let’s demonstrate using multiple years of AQI data
Multiple years of Tucson, AZ data
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2017.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2018.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2019.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2020.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2021.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2022.csv
/Users/gchism/Desktop/INFO-526-SU24/slides/07/data/tucson/ad_aqi_tracker_data-2023.csvReading multiple files
tuc <- read_csv(tuc_files, na = c(".", ""))
tuc <- tuc |>
janitor::clean_names() |>
mutate(
date = mdy(date),
good_aqi = if_else(aqi_value <= 50, 1, 0)
) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(cumsum_good_aqi = cumsum(good_aqi), .after = aqi_value)
tuc# A tibble: 2,392 × 13
date aqi_value cumsum_good_aqi main_pollutant site_name
<date> <dbl> <dbl> <chr> <chr>
1 2017-01-01 38 1 Ozone SAGUARO PA…
2 2017-01-02 40 2 Ozone SAGUARO PA…
3 2017-01-03 38 3 Ozone SAGUARO PA…
4 2017-01-04 38 4 Ozone SAGUARO PA…
5 2017-01-05 32 5 Ozone SAGUARO PA…
# ℹ 2,387 more rows
# ℹ 8 more variables: site_id <chr>, source <chr>,
# x20_year_high_2000_2019 <dbl>, x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>, good_aqi <dbl>Plot trend since 2016

tuc |> ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) + geom_smooth(method = "lm", color = "pink") + geom_line() + scale_x_date( expand = expansion(mult = 0.02), date_labels = "%Y" ) + labs( x = NULL, y = "Number of days", title = "Cumulative number of good AQI days (AQI < 50)", subtitle = "Tucson, AZ", caption = "\nSource: EPA Daily Air Quality Tracker" ) + theme(plot.title.position = "plot")tuc |> ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) + geom_smooth(method = "lm", color = "pink") + geom_line() + scale_x_date( expand = expansion(mult = 0.02), date_labels = "%Y" ) + labs( x = NULL, y = "Number of days", title = "Cumulative number of good AQI days (AQI < 50)", subtitle = "Tucson, AZ", caption = "\nSource: EPA Daily Air Quality Tracker" ) + theme(plot.title.position = "plot")
`geom_smooth()` using formula = 'y ~ x'Detrend
Step 1. Fit a simple linear regression
Detrend
Step 2. Augment the data with model results (using broom::augment())
# A tibble: 2,392 × 8
cumsum_good_aqi date .fitted .resid .hat .sigma
<dbl> <date> <dbl> <dbl> <dbl> <dbl>
1 1 2017-01-01 -0.574 1.57 0.00167 23.9
2 2 2017-01-02 0.0457 1.95 0.00167 23.9
3 3 2017-01-03 0.666 2.33 0.00167 23.9
4 4 2017-01-04 1.29 2.71 0.00166 23.9
5 5 2017-01-05 1.91 3.09 0.00166 23.9
# ℹ 2,387 more rows
# ℹ 2 more variables: .cooksd <dbl>, .std.resid <dbl>Detrend
Step 3. Divide the observed value of cumsum_good_aqi by the respective value in the long-term trend (i.e., .fitted)
# A tibble: 2,392 × 9
cumsum_good_aqi date .fitted ratio .resid .hat .sigma
<dbl> <date> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 2017-01-01 -0.574 -1.74 1.57 0.00167 23.9
2 2 2017-01-02 0.0457 43.7 1.95 0.00167 23.9
3 3 2017-01-03 0.666 4.51 2.33 0.00167 23.9
4 4 2017-01-04 1.29 3.11 2.71 0.00166 23.9
5 5 2017-01-05 1.91 2.62 3.09 0.00166 23.9
# ℹ 2,387 more rows
# ℹ 2 more variables: .cooksd <dbl>, .std.resid <dbl>Visualize detrended data

tuc_aug |>
ggplot(aes(x = date, y = ratio, group = 1)) +
geom_hline(yintercept = 1, color = "gray") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.1),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days\n(detrended)",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "Tucson, AZ",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Air Quality in Tucson
barely anything interesting happening!
let’s look at data from somewhere with a bit more “interesting” air quality data…
Read in multiple years of SF data
sf <- read_csv(sf_files, na = c(".", ""))
sf <- sf |>
janitor::clean_names() |>
mutate(
date = mdy(date),
good_aqi = if_else(aqi_value <= 50, 1, 0)
) |>
filter(!is.na(aqi_value)) |>
arrange(date) |>
mutate(cumsum_good_aqi = cumsum(good_aqi), .after = aqi_value)
sf# A tibble: 2,557 × 13
date aqi_value cumsum_good_aqi main_pollutant site_name
<date> <dbl> <dbl> <chr> <chr>
1 2016-01-01 32 1 PM2.5 Durham Arm…
2 2016-01-02 37 2 PM2.5 Durham Arm…
3 2016-01-03 45 3 PM2.5 Durham Arm…
4 2016-01-04 33 4 PM2.5 Durham Arm…
5 2016-01-05 27 5 PM2.5 Durham Arm…
# ℹ 2,552 more rows
# ℹ 8 more variables: site_id <chr>, source <chr>,
# x20_year_high_2000_2019 <dbl>, x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>, good_aqi <dbl>Plot trend since 2016

sf |>
ggplot(aes(x = date, y = cumsum_good_aqi, group = 1)) +
geom_smooth(method = "lm", color = "pink") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.07),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "San Francisco-Oakland-Hayward, CA",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")`geom_smooth()` using formula = 'y ~ x'Detrend
- Fit a simple linear regression
Visualize detrended data

sf_aug |>
ggplot(aes(x = date, y = ratio, group = 1)) +
geom_hline(yintercept = 1, color = "gray") +
geom_line() +
scale_x_date(
expand = expansion(mult = 0.07),
date_labels = "%Y"
) +
labs(
x = NULL, y = "Number of days\n(detrended)",
title = "Cumulative number of good AQI days (AQI < 50)",
subtitle = "San Francisco-Oakland-Hayward, CA",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(plot.title.position = "plot")Detrending
In step 2 we fit a very simple model
Depending on the complexity you’re trying to capture you might choose to fit a much more complex model
You can also decompose the trend into multiple trends, e.g. monthly, long-term, seasonal, etc.