Visualizing time series data I
Lecture 6
University of Arizona
INFO 526 - Summer 2024
HW 1 lessons
Highlights
Review HW 1 issues, and show me you reviewed them by closing the issue.
DO NOT hard code paths! Use the
herepackage to help with relative paths, if you need. e.g.,data <- read_csv(here("data", "data.csv"))Make sure that all packages are installed/loaded. I suggest the
pacmanworkflow we saw before:
HW 1 lessons learned cont…
- Start very early. No late work exceptions
- Ask your peers.
- Peers will likely have the answer
- Peers will likely get to the question before I will.
- Ask descriptive questions. See this page on asking effective questions.
- Please respect my work hours. I do not reply to messages after 5pm on work days and at all on weekends.
Setup
# load packages
library(countdown)
library(tidyverse)
library(janitor)
library(colorspace)
library(broom)
library(fs)
# set theme for ggplot2
ggplot2::theme_set(ggplot2::theme_minimal(base_size = 14))
# set width of code output
options(width = 65)
# set figure parameters for knitr
knitr::opts_chunk$set(
fig.width = 7, # 7" width
fig.asp = 0.618, # the golden ratio
fig.retina = 3, # dpi multiplier for displaying HTML output on retina
fig.align = "center", # center align figures
dpi = 300 # higher dpi, sharper image
)Working with dates
Air Quality Index
The AQI is the Environmental Protection Agency’s index for reporting air quality
Higher values of AQI indicate worse air quality

AQI levels
The previous graphic in tibble form, to be used later…
AQI data
Source: EPA’s Daily Air Quality Tracker
2016 - 2022 AQI (Ozone and PM2.5 combined) for Tucson, AZ core-based statistical area (CBSA), one file per year
2016 - 2022 AQI (Ozone and PM2.5 combined) for San Francisco-Oakland-Hayward, CA CBSA, one file per year
2022 Tucson, AZ
- Load data
Clean variable names
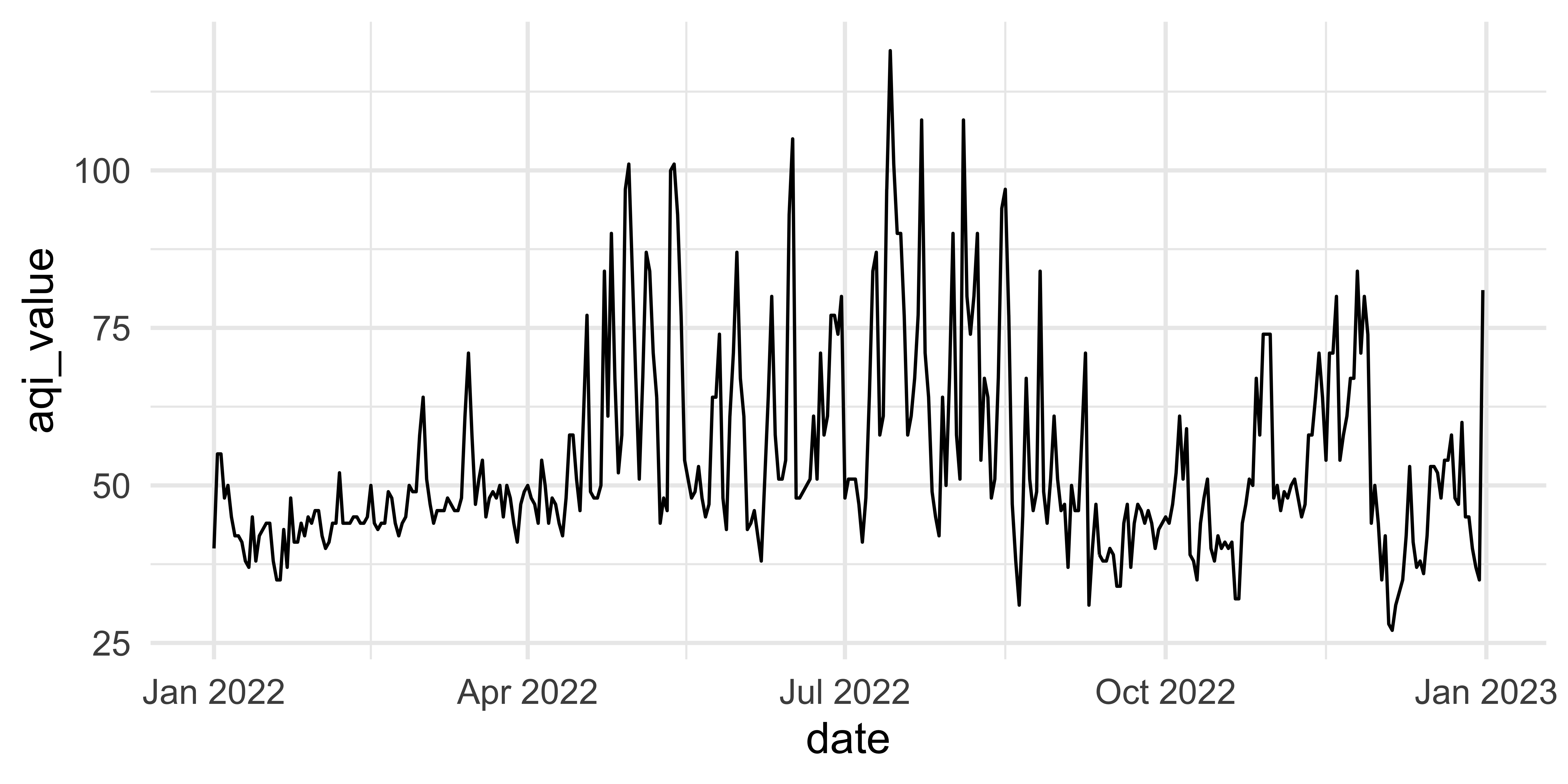
First look
This plot looks quite bizarre. What might be going on?
Peek at data
# A tibble: 365 × 4
date aqi_value site_name site_id
<chr> <dbl> <chr> <chr>
1 01/01/2022 40 GERONIMO 04-019-1113
2 01/02/2022 55 GERONIMO 04-019-1113
3 01/03/2022 55 GERONIMO 04-019-1113
4 01/04/2022 48 GERONIMO 04-019-1113
5 01/05/2022 50 GERONIMO 04-019-1113
6 01/06/2022 45 GERONIMO 04-019-1113
7 01/07/2022 42 SAGUARO PARK 04-019-0021
8 01/08/2022 42 SAGUARO PARK 04-019-0021
9 01/09/2022 41 GERONIMO 04-019-1113
10 01/10/2022 38 FAIRGROUNDS 04-019-1020
# ℹ 355 more rowsTransforming date
Using lubridate::mdy():
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source
<date> <dbl> <chr> <chr> <chr> <chr>
1 2022-01-01 40 PM2.5 GERONIMO 04-019… AQS
2 2022-01-02 55 PM2.5 GERONIMO 04-019… AQS
3 2022-01-03 55 PM2.5 GERONIMO 04-019… AQS
4 2022-01-04 48 PM2.5 GERONIMO 04-019… AQS
5 2022-01-05 50 PM2.5 GERONIMO 04-019… AQS
6 2022-01-06 45 PM2.5 GERONIMO 04-019… AQS
7 2022-01-07 42 Ozone SAGUARO PA… 04-019… AQS
8 2022-01-08 42 Ozone SAGUARO PA… 04-019… AQS
9 2022-01-09 41 PM2.5 GERONIMO 04-019… AQS
10 2022-01-10 38 Ozone FAIRGROUNDS 04-019… AQS
# ℹ 355 more rows
# ℹ 5 more variables: x20_year_high_2000_2019 <dbl>,
# x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
# date_of_20_year_low <chr>Investigating AQI values
- Take a peek at distinct values of AQI
[1] 40 55 48 50 45 42 41 38 37 43 44 35 46 52 49
[16] 58 64 51 47 61 71 54 77 84 90 67 97 101 87 100
[31] 93 53 74 80 105 119 108 94 31 39 34 59 32 28 27
[46] 33 36 60 81"."likely indicatesNA, and it’s causing the entire column to be read in as characters
Rewind, and start over
Rows: 365
Columns: 11
$ Date <chr> "01/01/2022", "01/02/2022"…
$ `AQI Value` <dbl> 40, 55, 55, 48, 50, 45, 42…
$ `Main Pollutant` <chr> "PM2.5", "PM2.5", "PM2.5",…
$ `Site Name` <chr> "GERONIMO", "GERONIMO", "G…
$ `Site ID` <chr> "04-019-1113", "04-019-111…
$ Source <chr> "AQS", "AQS", "AQS", "AQS"…
$ `20-year High (2000-2019)` <dbl> 115, 57, 67, 55, 52, 55, 5…
$ `20-year Low (2000-2019)` <dbl> 35, 31, 29, 27, 28, 25, 31…
$ `5-year Average (2015-2019)` <dbl> 62.2, 43.2, 43.6, 40.4, 36…
$ `Date of 20-year High` <chr> "01/01/2018", "01/02/2015"…
$ `Date of 20-year Low` <chr> "01/01/2001", "01/02/2016"…Data cleaning
# A tibble: 365 × 11
date aqi_value main_pollutant site_name site_id source
<date> <dbl> <chr> <chr> <chr> <chr>
1 2022-01-01 40 PM2.5 GERONIMO 04-019… AQS
2 2022-01-02 55 PM2.5 GERONIMO 04-019… AQS
3 2022-01-03 55 PM2.5 GERONIMO 04-019… AQS
4 2022-01-04 48 PM2.5 GERONIMO 04-019… AQS
5 2022-01-05 50 PM2.5 GERONIMO 04-019… AQS
6 2022-01-06 45 PM2.5 GERONIMO 04-019… AQS
7 2022-01-07 42 Ozone SAGUARO PA… 04-019… AQS
8 2022-01-08 42 Ozone SAGUARO PA… 04-019… AQS
9 2022-01-09 41 PM2.5 GERONIMO 04-019… AQS
10 2022-01-10 38 Ozone FAIRGROUNDS 04-019… AQS
# ℹ 355 more rows
# ℹ 5 more variables: x20_year_high_2000_2019 <dbl>,
# x20_year_low_2000_2019 <dbl>,
# x5_year_average_2015_2019 <dbl>, date_of_20_year_high <chr>,
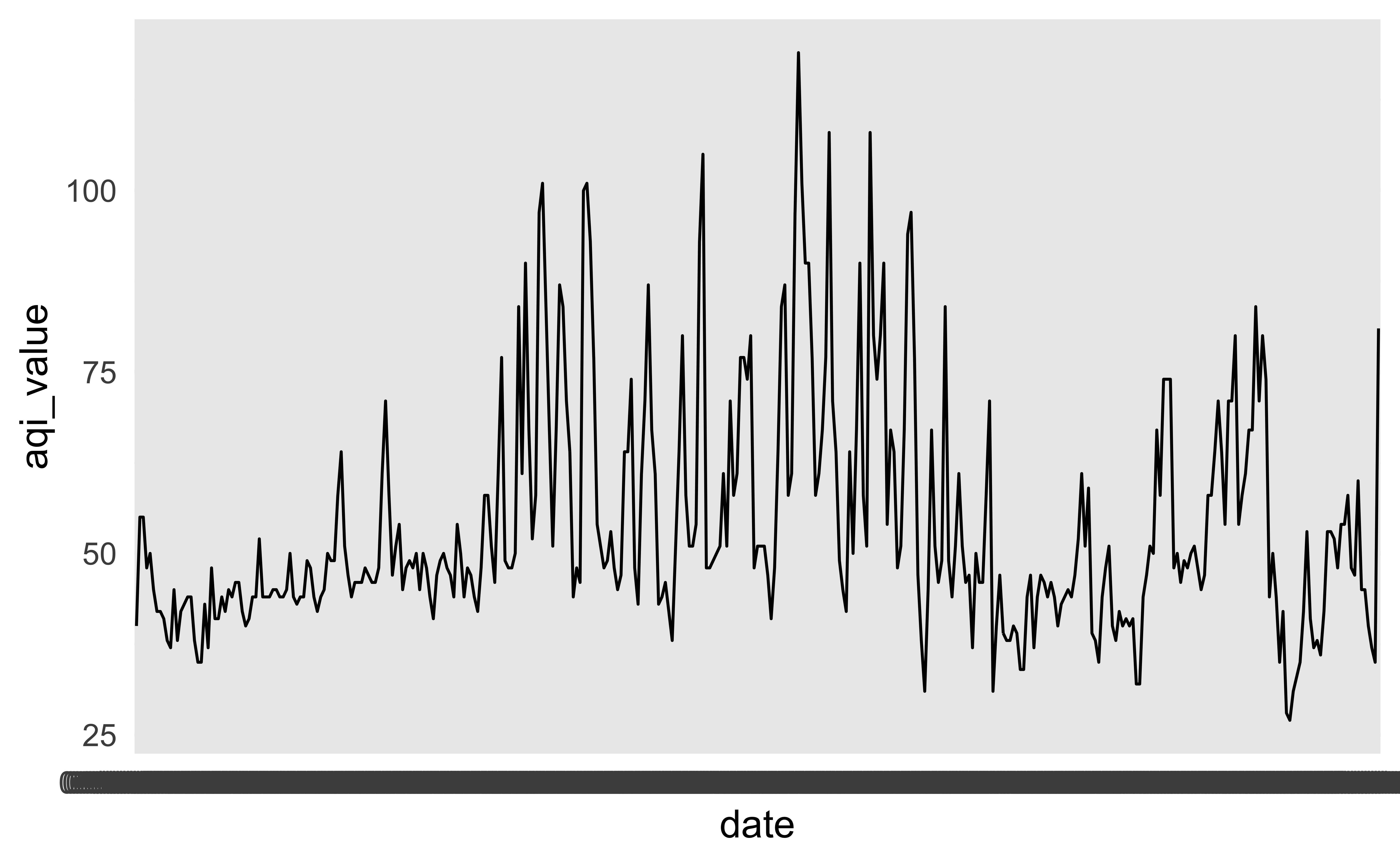
# date_of_20_year_low <chr>Another look
How would you improve this visualization?
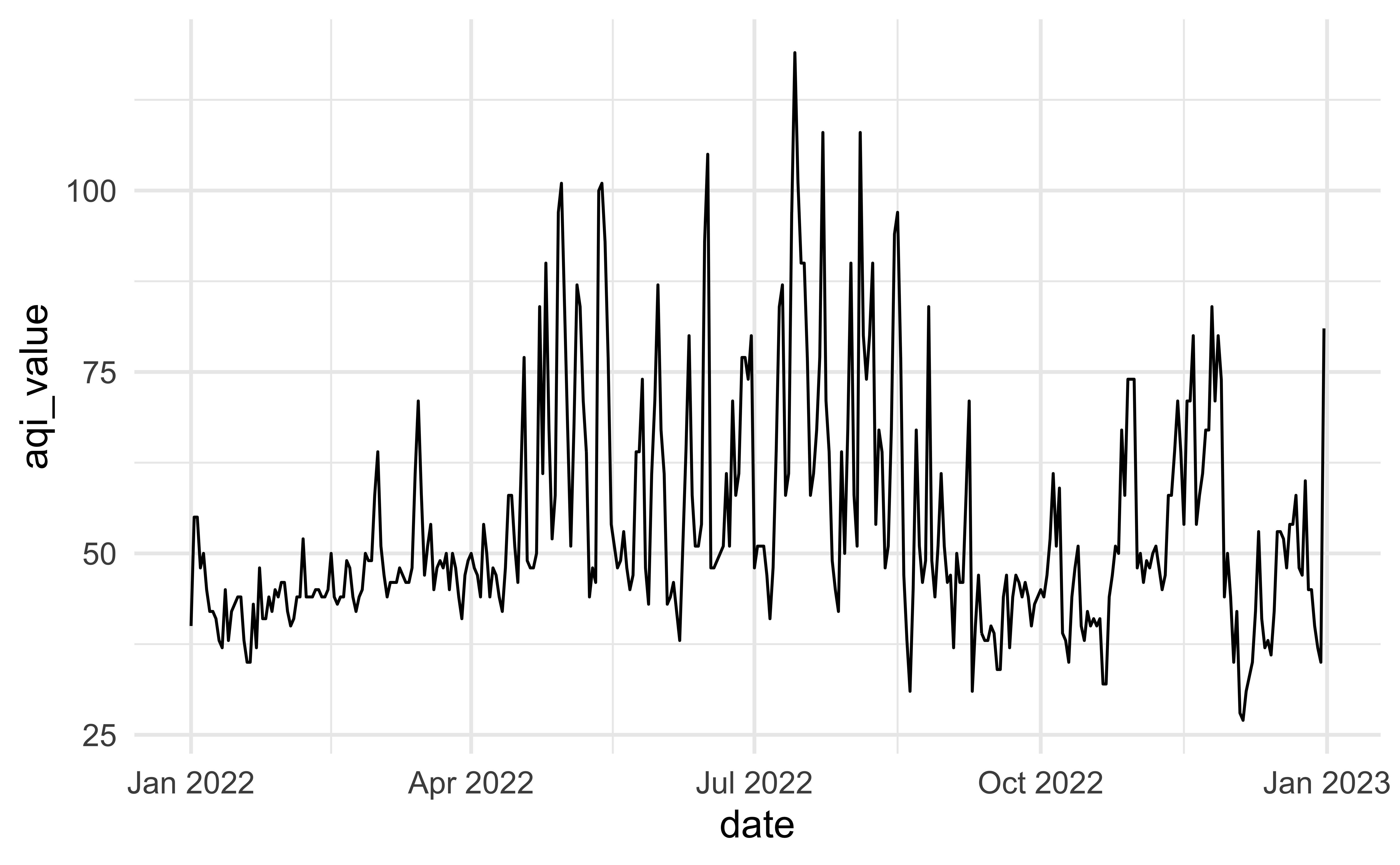
Visualizing Tucson AQI
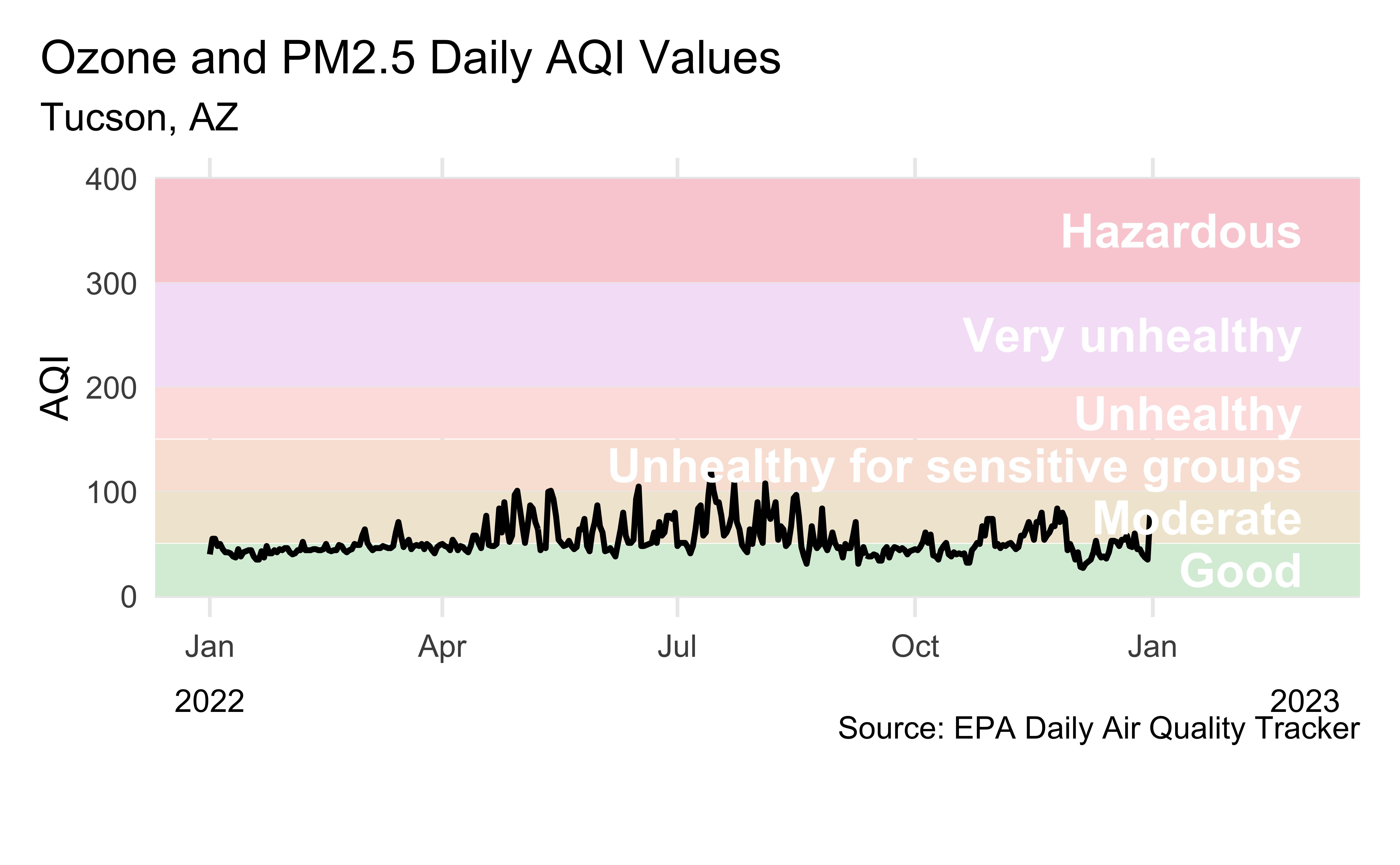
Livecoding
Setup
aqi_levels <- tribble(
~aqi_min, ~aqi_max, ~color, ~level,
0, 50, "#D8EEDA", "Good",
51, 100, "#F1E7D4", "Moderate",
101, 150, "#F8E4D8", "Unhealthy for sensitive groups",
151, 200, "#FEE2E1", "Unhealthy",
201, 300, "#F4E3F7", "Very unhealthy",
301, 400, "#F9D0D4", "Hazardous"
)
tuc_2022 <- read_csv("https://raw.githubusercontent.com/INFO-526-SU24/INFO-526-SU24/main/slides/06/data/tucson/ad_aqi_tracker_data-2022.csv",
na = c(".", ""))
tuc_2022 <- tuc_2022 |>
janitor::clean_names() |>
mutate(date = mdy(date))Reveal below for code developed during live coding session.
Code
aqi_levels <- aqi_levels |>
mutate(aqi_mid = ((aqi_min + aqi_max) / 2))
tuc_2022 |>
filter(!is.na(aqi_value)) |>
ggplot(aes(x = date, y = aqi_value, group = 1)) +
geom_rect(
data = aqi_levels,
aes(
ymin = aqi_min, ymax = aqi_max,
xmin = as.Date(-Inf), xmax = as.Date(Inf),
fill = color, y = NULL, x = NULL
)
) +
geom_line(linewidth = 1) +
scale_fill_identity() +
scale_x_date(
name = NULL, date_labels = "%b",
limits = c(ymd("2022-01-01"), ymd("2023-03-01"))
) +
geom_text(
data = aqi_levels,
aes(x = ymd("2023-02-28"), y = aqi_mid, label = level),
hjust = 1, size = 6, fontface = "bold", color = "white"
) +
annotate(
geom = "text",
x = c(ymd("2022-01-01"), ymd("2023-03-01")), y = -100,
label = c("2022", "2023"), size = 4
) +
coord_cartesian(clip = "off", ylim = c(0, 400)) +
labs(
x = NULL, y = "AQI",
title = "Ozone and PM2.5 Daily AQI Values",
subtitle = "Tucson, AZ",
caption = "\nSource: EPA Daily Air Quality Tracker"
) +
theme(
plot.title.position = "plot",
panel.grid.minor.y = element_blank(),
panel.grid.minor.x = element_blank(),
plot.margin = unit(c(1, 1, 3, 1), "lines")
)